Molecular and Genetic Characterization of the Notch Signaling Network
A major incentive in embarking on the very large project of defining the protein-interaction map of Drosophilahas been our desire to gain some insight into the underlying molecular parameters that govern the extremely complex gene circuitry that modulates Notch activity. We are thus very interested in correlating genetic interactions revealed by our functional Notch genetic screens with protein-protein interactions revealed by the proteome map. We are focusing our attention on the gentic interactors of approximately 150 core genes that have been associated with Notch signaling. We are taking advantage of the Exelixis mutant collection (see below) to verify and further explore the biological and mechanistic relevance of these interactions in vivo. It is our hope that this systems-level study will provide insights into the Notch genetic circuitry and reveal high-level properties that govern this pathway.
Ho, DM, Guruharsha, KG, and Artavanis-Tsakonas, S. (2018). The Notch interactome: Complexity in signaling circuitry. In Advances in Experimental Medicine and Biology, (Springer New York LLC), pp. 125–140.

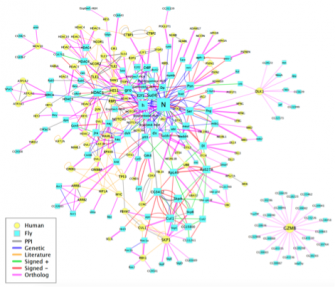

 The fundamental, conserved Notch signaling pathway controls a vast range of biological processes during development and homeostasis, including proliferation, differentiation, cell death, and stem cell maintenance. Notch pathway perturbations have also been implicated in a wide range of cancers. The interaction of Notch with other proteins and pathways in the cell plays a large role in determining the ultimate biological output of Notch signaling. To identify potential partners of Notch that have consequences for hyperplasia and oncogenesis, we have carried out an unbiased genome-wide screen to identify genes that affect Notch-induced proliferation, using the fruit fly Drosophila melanogaster. In addition to enhancing or suppressing a Notch-induced hyperproliferation phenotype, several of these interactions also caused a de novoectopic eye phenotype reminiscent of metastatic
The fundamental, conserved Notch signaling pathway controls a vast range of biological processes during development and homeostasis, including proliferation, differentiation, cell death, and stem cell maintenance. Notch pathway perturbations have also been implicated in a wide range of cancers. The interaction of Notch with other proteins and pathways in the cell plays a large role in determining the ultimate biological output of Notch signaling. To identify potential partners of Notch that have consequences for hyperplasia and oncogenesis, we have carried out an unbiased genome-wide screen to identify genes that affect Notch-induced proliferation, using the fruit fly Drosophila melanogaster. In addition to enhancing or suppressing a Notch-induced hyperproliferation phenotype, several of these interactions also caused a de novoectopic eye phenotype reminiscent of metastatic 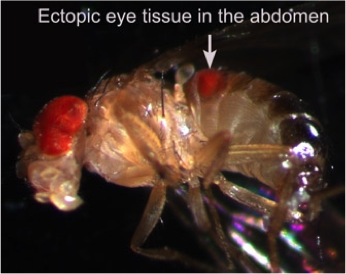 tissue. After screening approximately 16,000 mutant lines, we have identified 360 unique Notch interactors, 206 of which have clear human orthologs.
tissue. After screening approximately 16,000 mutant lines, we have identified 360 unique Notch interactors, 206 of which have clear human orthologs.